Probe Molecules¶
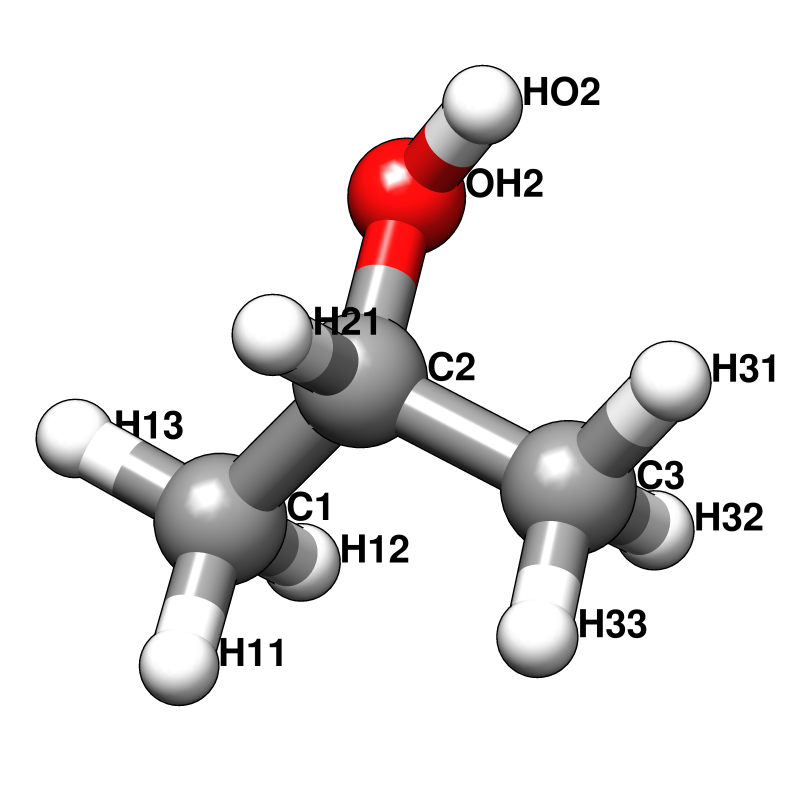
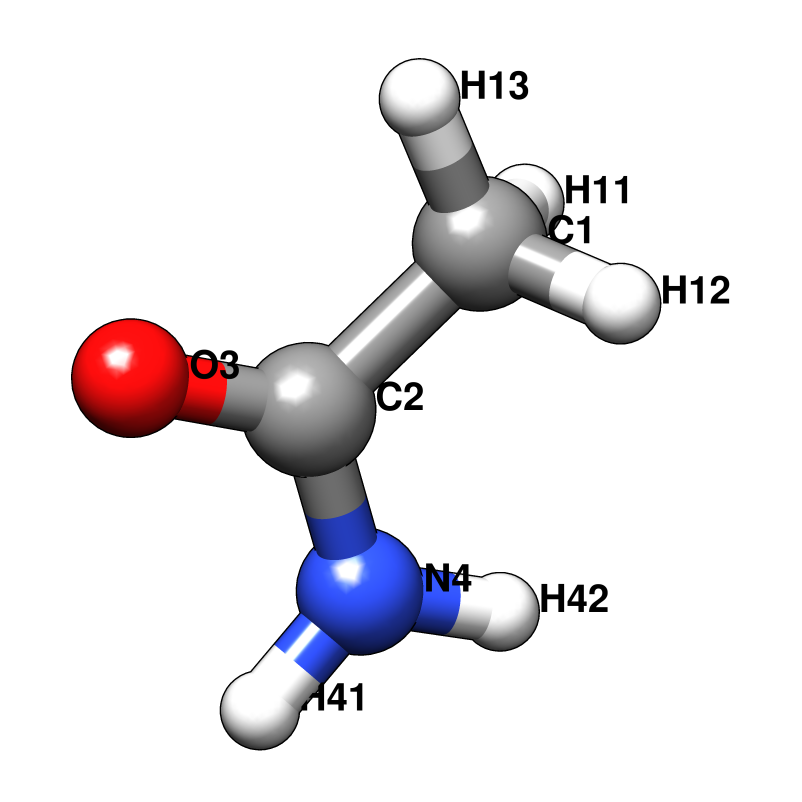
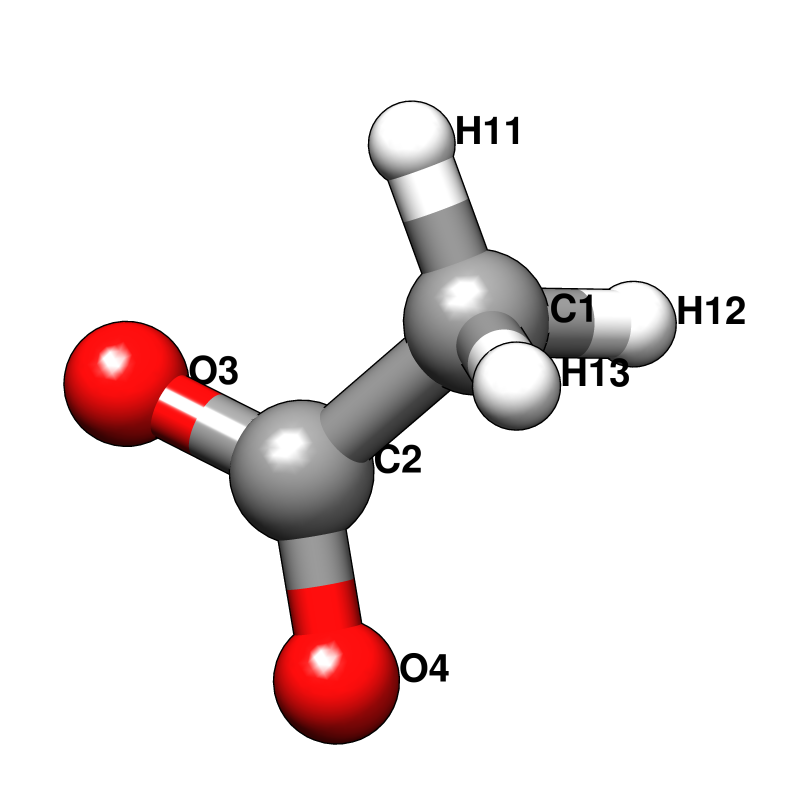
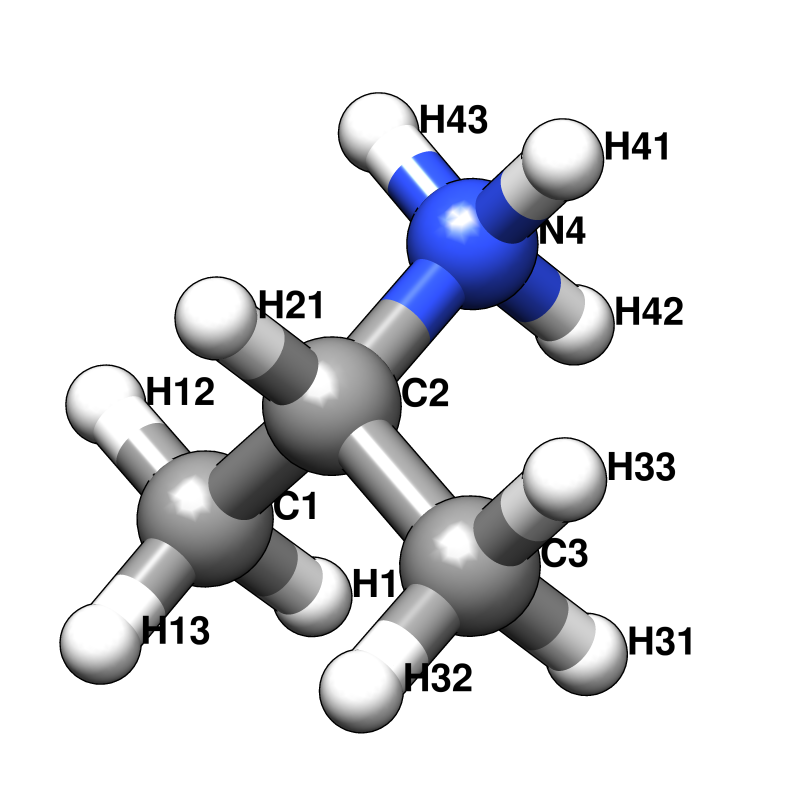
Probe molecules and their fractional composition, shown below, were selected based on analysis of FDA approved drugs [AB12]. The best composition for a specific system may, however, depend on the surface properties of the target. For a protein with a highly charged surface, increasing the proportion of acetate and isopropylamine might perform better in identifying ligandable sites.
Among these, isobutane is not included in the default configuration due to its high propensity to aggregate. If it is included, it should have a small fraction.
Topology & Parameter¶
Probe atom types and parameters are based on protein side-chains described in CHARMM force field. Below topology and parameter files distributed with the plugin are shown:
* -------------------------------------------------------------------------- *
* Topology file for probes used in druggability calculations *
* 31 December, 2009 *
* -------------------------------------------------------------------------- *
*
36 1
! -------------------------------------------------------------------------- !
! ATOM NAME SERIAL NO. SLOTS !
! RESERVED FOR THE ATOMS !
! Hydrogens 1 -- 39 !
! Carbons 40 -- 99 !
! Nitrogens 100 -- 149 !
! Oxygens 150 -- 189 !
! Sulphurs 190 -- 209 !
! Halogens 210 -- 249 !
! Miscellaneous 250 -- --- !
! -------------------------------------------------------------------------- !
!hydrogens
MASS 1 H 1.00800 H ! polar H
MASS 2 HC 1.00800 H ! N-ter H
MASS 3 HA 1.00800 H ! nonpolar H
MASS 4 HT 1.00800 H ! TIPS3P WATER HYDROGEN
MASS 32 CC 12.01100 C ! carbonyl C, asn,asp,gln,glu,cter,ct2
MASS 55 NH2 14.00700 N ! amide nitrogen
MASS 56 NH3 14.00700 N ! ammonium nitrogen
MASS 70 O 15.99900 O ! carbonyl oxygen
MASS 72 OC 15.99900 O ! carboxylate oxygen
MASS 75 OT 15.99940 O ! TIPS3P WATER OXYGEN
MASS 22 CT1 12.01100 C ! aliphatic sp3 C for CH
MASS 24 CT3 12.01100 C ! aliphatic sp3 C for CH3
MASS 73 OH1 15.99900 O ! hydroxyl oxygen
DEFA FIRS NONE LAST NONE
AUTO ANGLES DIHE
RESI TIP3 0.000 ! tip3p water model, generate using noangle nodihedral
GROUP
ATOM OH2 OT -0.834
ATOM H1 HT 0.417
ATOM H2 HT 0.417
BOND OH2 H1 OH2 H2 !H1 H2 ! the last bond is needed for shake
!ANGLE H1 OH2 H2 ! required
ACCEPTOR OH2
PATCHING FIRS NONE LAST NONE
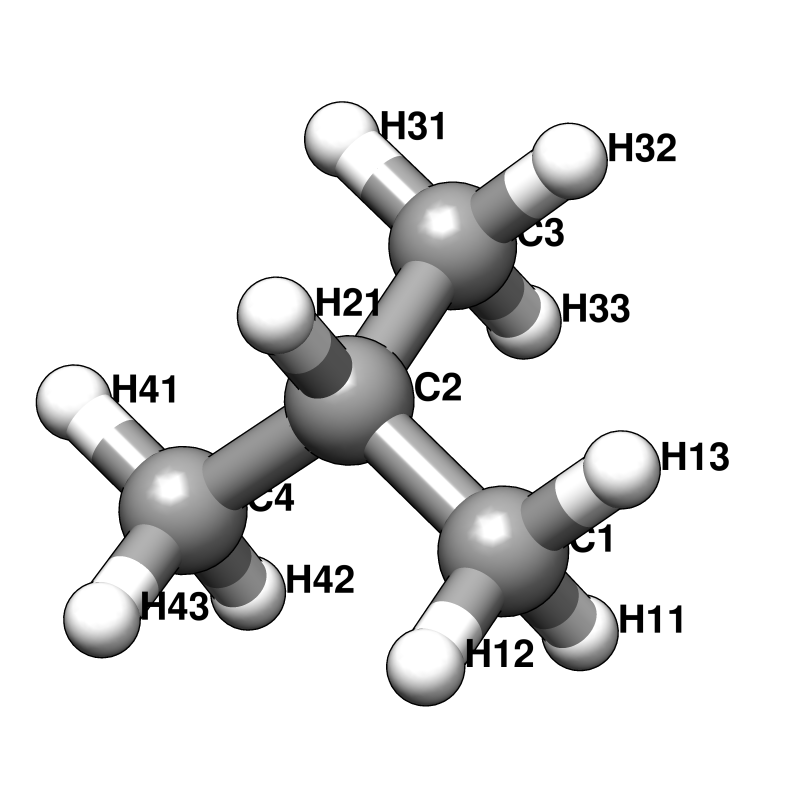
RESI IPRO 0.000 ! ATOM TYPES FROM THR
GROUP
ATOM C2 CT1 0.181 ! H12 H13 H33 H32
ATOM H21 HA 0.049 ! \ / \ /
GROUP ! H11--C1 C3--H31
ATOM C1 CT3 -0.147 ! \ /
ATOM H11 HA 0.049 ! C2
ATOM H12 HA 0.049 ! / \
ATOM H13 HA 0.049 ! OH2 H21
GROUP ! |
ATOM C3 CT3 -0.147 ! HO2
ATOM H31 HA 0.049
ATOM H32 HA 0.049
ATOM H33 HA 0.049
GROUP
ATOM OH2 OH1 -0.660
ATOM HO2 H 0.430
BOND C2 C1 C2 C3 C2 OH2
BOND C1 H11 C1 H12 C1 H13
BOND C2 H21
BOND C3 H31 C3 H32 C3 H33
BOND OH2 HO2
DONOR HO2 OH2
ACCEPTOR OH2
RESI IBUT 0.000 ! ISOBUTANE
GROUP
ATOM C2 CT1 -0.049 ! H12 H13 H33 H32
ATOM H21 HA 0.049 ! \ / \ /
GROUP ! H11--C1 C3--H31
ATOM C1 CT3 -0.147 ! \ /
ATOM H11 HA 0.049 ! C2--H21
ATOM H12 HA 0.049 ! /
ATOM H13 HA 0.049 ! H41--C4--H43
GROUP ! |
ATOM C3 CT3 -0.147 ! H42
ATOM H31 HA 0.049
ATOM H32 HA 0.049
ATOM H33 HA 0.049
GROUP
ATOM C4 CT3 -0.147
ATOM H41 HA 0.049
ATOM H42 HA 0.049
ATOM H43 HA 0.049
BOND C2 C1 C2 C3 C2 C4
BOND C1 H11 C1 H12 C1 H13
BOND C2 H21
BOND C3 H31 C3 H32 C3 H33
BOND C4 H41 C4 H42 C4 H43
IC C2 H13 *C1 H11 1.5472 117.4600 120.9800 107.1700 1.1145
IC C2 H13 *C1 H12 1.5472 117.4600 -124.6700 108.9800 1.1126
IC H13 C1 C2 C3 1.5543 117.4600 180.0000 110.4800 1.5361
IC C3 C1 *C2 C4 1.5361 110.4800 120.0000 112.5700 1.5360
IC C3 C4 *C2 H21 1.5361 110.2600 120.0000 108.0200 1.1168
IC C1 C2 C3 H31 1.5472 110.4800 177.3300 110.5400 1.1111
IC H31 C2 *C3 H32 1.1111 110.5400 119.9600 110.6200 1.1112
IC H31 C2 *C3 H33 1.1111 110.5400 -119.8500 110.6900 1.1108
IC C1 C2 C4 H41 1.5472 112.5700 178.9600 110.3200 1.1116
IC H41 C2 *C4 H42 1.1116 110.3200 119.7100 111.6900 1.1086
IC H41 C2 *C4 H43 1.1116 110.3200 -119.6100 110.4900 1.1115
RESI IPAM 1.000 ! ISOPROPYLAMINE, ATOM TYPES FROM LYS
GROUP
ATOM C2 CT1 0.252 ! H12 H13 H33 H32
ATOM H21 HA 0.049 ! \ / \ /
GROUP ! H11--C1 C3--H31
ATOM C1 CT3 -0.147 ! \ /
ATOM H11 HA 0.049 ! C2--H21
ATOM H12 HA 0.049 ! /
ATOM H13 HA 0.049 ! H41--N4--H43
GROUP ! |
ATOM C3 CT3 -0.147 ! H42
ATOM H31 HA 0.049
ATOM H32 HA 0.049
ATOM H33 HA 0.049
GROUP
ATOM N4 NH3 -0.300
ATOM H41 HC 0.333
ATOM H42 HC 0.333
ATOM H43 HC 0.333
BOND C2 C1 C2 C3 C2 N4
BOND C1 H11 C1 H12 C1 H13
BOND C2 H21
BOND C3 H31 C3 H32 C3 H33
BOND N4 H41 N4 H42 N4 H43
IC C2 H13 *C1 H11 1.5472 117.4600 120.9800 107.1700 1.1145
IC C2 H13 *C1 H12 1.5472 117.4600 -124.6700 108.9800 1.1126
IC H13 C1 C2 C3 1.5543 117.4600 180.0000 110.4800 1.5361
IC C3 C1 *C2 N4 1.5361 110.4800 120.0000 112.5700 1.5360
IC C3 N4 *C2 H21 1.5361 110.2600 120.0000 108.0200 1.1168
IC C1 C2 C3 H31 1.5472 110.4800 177.3300 110.5400 1.1111
IC H31 C2 *C3 H32 1.1111 110.5400 119.9600 110.6200 1.1112
IC H31 C2 *C3 H33 1.1111 110.5400 -119.8500 110.6900 1.1108
IC C1 C2 N4 H41 1.5472 112.5700 178.9600 110.3200 1.1116
IC H41 C2 *N4 H42 1.1116 110.3200 119.7100 111.6900 1.1086
IC H41 C2 *N4 H43 1.1116 110.3200 -119.6100 110.4900 1.1115
RESI ACET -1.000 ! ACETATE, ATOM TYPES FROM GLU
GROUP
ATOM C2 CC 0.520 ! H11 O4
ATOM O3 OC -0.760 ! | //
ATOM O4 OC -0.760 ! H12--C1--C2
GROUP ! | \
ATOM C1 CT3 -0.147 ! H13 O3 (-)
ATOM H11 HA 0.049
ATOM H12 HA 0.049
ATOM H13 HA 0.049
BOND C2 C1 C2 O3 C2 O4
BOND C1 H11 C1 H12 C1 H13
IMPR C2 C1 O3 O4
IC C2 H13 *CB H11 1.5218 112.6000 119.2200 109.2300 1.1086
IC C2 H13 *CB H12 1.5218 112.6000 -121.6100 110.6400 1.1080
IC H13 CB C2 O4 1.5619 112.6000 180.0000 117.9900 1.2565
IC O4 CB *C2 O3 1.2565 117.9900 -170.2300 117.7000 1.2541
RESI ACAM 0.000 ! ACETAMIDE, ATOM TYPES FROM GLN
GROUP
ATOM C2 CC 0.550 ! H11 O3 H41
ATOM O3 O -0.550 ! | || /
GROUP ! H12--C1--C2--N4
ATOM C1 CT3 -0.147 ! | \
ATOM H11 HA 0.049 ! H13 H42
ATOM H12 HA 0.049
ATOM H13 HA 0.049
GROUP
ATOM N4 NH2 -0.620
ATOM H41 H 0.320
ATOM H42 H 0.300
BOND C2 C1 C2 O3 C2 N4
BOND C1 H11 C1 H12 C1 H13
BOND N4 H41 N4 H42
IMPR C2 N4 C1 O3 C2 C1 N4 O3
IMPR N4 C2 H41 H42 N4 C2 H42 H41
DONOR H41 N4
DONOR H42 N4
ACCEPTOR O3 C2
IC C2 H13 *CB H11 1.5319 114.3000 119.1700 107.8200 1.1120
IC C2 H13 *CB H12 1.5319 114.3000 -123.7400 110.3400 1.1091
IC H13 CB C2 O3 1.5627 114.3000 180.0000 122.5600 1.2323
IC O3 CB *C2 N4 1.2323 122.5600 -179.1900 116.1500 1.3521
IC CB C2 N4 H41 1.5319 116.1500 -179.2600 117.3500 0.9963
IC H41 C2 *N4 H42 0.9963 117.3500 178.0200 120.0500 0.9951
END
*>>>>>>>>>> Parameter file for druggability calculations <<<<<<<<<<<<<
*>>>>>>>>>>>>>>>>>>>>>>>> 31 December, 2009 <<<<<<<<<<<<<<<<<<<<<<<<<<<<
*
! references
!
!PROTEINS
!
!MacKerell, A.D., Jr,. Feig, M., Brooks, C.L., III, Extending the
!treatment of backbone energetics in protein force fields: limitations
!of gas-phase quantum mechanics in reproducing protein conformational
!distributions in molecular dynamics simulations, Journal of
!Computational Chemistry, 25: 1400-1415, 2004.
!
!MacKerell, Jr., A. D.; Bashford, D.; Bellott, M.; Dunbrack Jr., R.L.;
!Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.;
!Joseph-McCarthy, D.; Kuchnir, L.; Kuczera, K.; Lau, F.T.K.; Mattos,
!C.; Michnick, S.; Ngo, T.; Nguyen, D.T.; Prodhom, B.; Reiher, III,
!W.E.; Roux, B.; Schlenkrich, M.; Smith, J.C.; Stote, R.; Straub, J.;
!Watanabe, M.; Wiorkiewicz-Kuczera, J.; Yin, D.; Karplus, M. All-atom
!empirical potential for molecular modeling and dynamics Studies of
!proteins. Journal of Physical Chemistry B, 1998, 102, 3586-3616.
!
!
BONDS
!
!V(bond) = Kb(b - b0)**2
!
!Kb: kcal/mole/A**2
!b0: A
!
!atom type Kb b0
!
ANGLES
!
!V(angle) = Ktheta(Theta - Theta0)**2
!
!V(Urey-Bradley) = Kub(S - S0)**2
!
!Ktheta: kcal/mole/rad**2
!Theta0: degrees
!Kub: kcal/mole/A**2 (Urey-Bradley)
!S0: A
!
!atom types Ktheta Theta0 Kub S0
!
NH3 CT1 HA 45.000 107.50 35.00 2.10100 ! ALLOW ALI POL
! new stretch and bend; methylammonium (KK 03/10/92)
! NH3 CT2 HA
DIHEDRALS
!
!V(dihedral) = Kchi(1 + cos(n(chi) - delta))
!
!Kchi: kcal/mole
!n: multiplicity
!delta: degrees
!
!atom types Kchi n delta
!
!Neutral N terminus
IMPROPER
!
!V(improper) = Kpsi(psi - psi0)**2
!
!Kpsi: kcal/mole/rad**2
!psi0: degrees
!note that the second column of numbers (0) is ignored
!
!atom types Kpsi psi0
!
END