Introduction¶
Druggability Suite is a VMD plugin GUI and a Python module developed for setup and analysis of simulations described in [AB12].
Installation¶
- VMD 1.9.1 or later is required for using GUI. NAMD is required for running druggability simulations. Following are required for performing druggability analysis calculations:
- Download one of the following archive files:
Extract contents of the archive and copy
druguifolder to VMD TCL plugins directory, i.e.$VMDDIR/plugins/noarch/tcl/.Then, insert following line into
$VMDDIR/scripts/vmd/loadplugins.tclat line 186:vmd_install_extension drugui drugui_tk "Modeling/DruGUI"
If you are not sure where VMD directory is located, run vmd, and type the following command line in the VMD console:
global env; puts $env(VMDDIR)
DruGUI Plugin¶
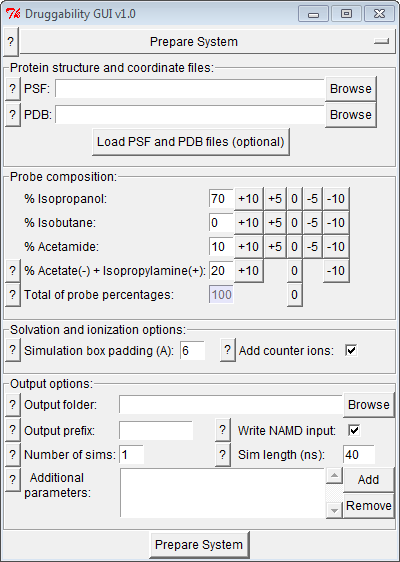
Druggability Suite GUI (DruGUI) plugin, shown below, has five panels to streamline setup, analysis, and visualization of druggability simulations:
The rest of the tutorial will show you how to use these panels, and described required inputs and outputs from different analysis steps.
Tutorial Files¶
Files in the following archives can be used to follow this tutorial:
Here is a list of these files:
3.5K mdm2_output.log
592K mdm2_output.pdb
1003K mdm2_output.psf
254 mdm2_output.xsc
112K mdm2.pdb
335K mdm2.psf
3.9K sample_1t4e_inhibitor.pdb
2.2M sample_ACAM.dx
2.2M sample_ACET.dx
4.5K sample_all_hotspots.pdb
55K sample_heavyatoms.pdb
2.2M sample_IPAM.dx
2.4M sample_IPRO.dx
5.7K sample.log
1.3K sample_site_1.pdb
553 sample_site_1_soln_1.pdb
553 sample_site_1_soln_2.pdb
553 sample_site_1_soln_3.pdb
How to Cite¶
If you benefited from Druggability Suite in your research, please cite the following paper:
| [AB12] | Bakan A, Nevins N, Lakdawala AS, Bahar I Druggability Assessment of Allosteric Proteins by Dynamics Simulations in the Presence of Probe Molecules J Chem Theory Comput 2012 8(7):2435-2447 |