Building Biomolecules¶
Some PDB files contain coordinates for a monomer of a functional/biological
multimer (biomolecule). ProDy offers functions to build structures of
biomolecules using the header data from the PDB file. We will use PDB file
that contains the coordinates for a monomer of a biological
multimeric protein and the transformations in the header section to
generate the multimer coordinates. Output will be an AtomGroup
instance that contains the multimer coordinates.
We start by importing everything from the ProDy package:
In [1]: from prody import *
In [2]: from pylab import *
In [3]: ion()
Build a Multimer¶
Let’s build the dimeric form of 3enl of enolase:
In [4]: monomer, header = parsePDB('3enl', header=True)
In [5]: monomer
Out[5]: <AtomGroup: 3enl (3647 atoms)>
Note that we passed header=True argument to parse header data in addition
to coordinates.
In [6]: showProtein(monomer);
In [7]: legend();
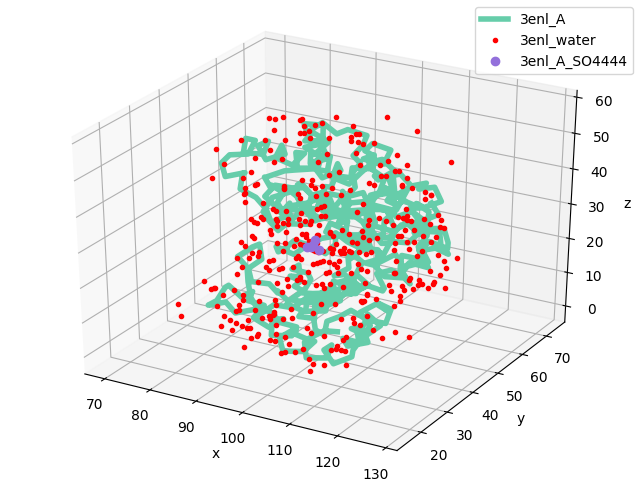
Let’s get the dimer coordinates using buildBiomolecules() function:
In [8]: dimer = buildBiomolecules(header, monomer)
In [9]: dimer
Out[9]: <AtomGroup: 3enl biomolecule 1 (7294 atoms)>
This function takes biomolecular tarnsformations from the header dictionary
(item with key 'biomoltrans') and applies them to the
monomer.
In [10]: showProtein(dimer);
In [11]: legend();
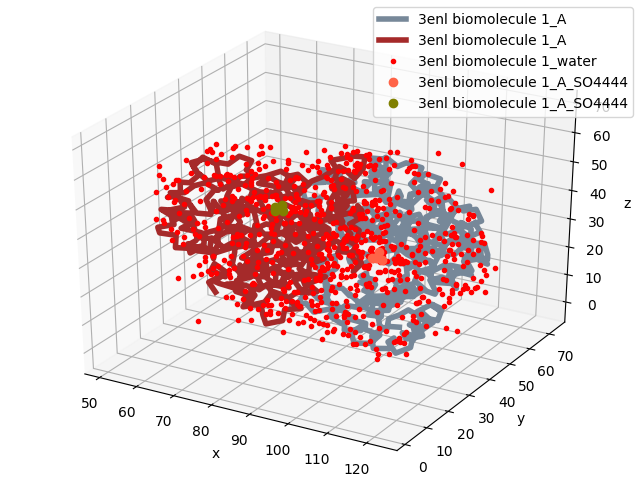
The dimer object now has two chains:
In [12]: list(dimer.iterChains())
Out[12]:
[<Chain: A from Segment A from 3enl biomolecule 1 (790 residues, 3647 atoms)>,
<Chain: A from Segment B from 3enl biomolecule 1 (790 residues, 3647 atoms)>]
Build a Tetramer¶
Let’s build the tetrameric form of 1k4c of KcsA_potassium_channel:
In [13]: monomer, header = parsePDB('1k4c', header=True)
In [14]: monomer
Out[14]: <AtomGroup: 1k4c (4534 atoms)>
In [15]: showProtein(monomer);
In [16]: legend();
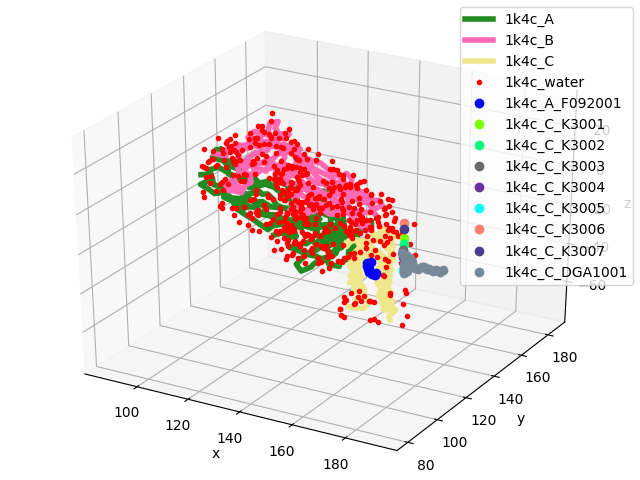
Note that we do not want to replicate potassium ions, so we will exclude them:
In [17]: potassium = monomer.name_K
In [18]: potassium
Out[18]: <Selection: 'name K' from 1k4c (7 atoms)>
In [19]: without_K = ~ potassium
In [20]: without_K
Out[20]: <Selection: 'not (name K)' from 1k4c (4527 atoms)>
In [21]: tetramer = buildBiomolecules(header, without_K)
In [22]: tetramer
Out[22]: <AtomGroup: 1k4c Selection 'not (name K)' biomolecule 1 (18108 atoms)>
Now, let’s append potassium ions to the tetramer:
In [23]: potassium.setChids('K')
In [24]: kcsa = tetramer + potassium.copy()
In [25]: kcsa.setTitle('KcsA')
Here is a view of the tetramer:
In [26]: showProtein(kcsa);
In [27]: legend();
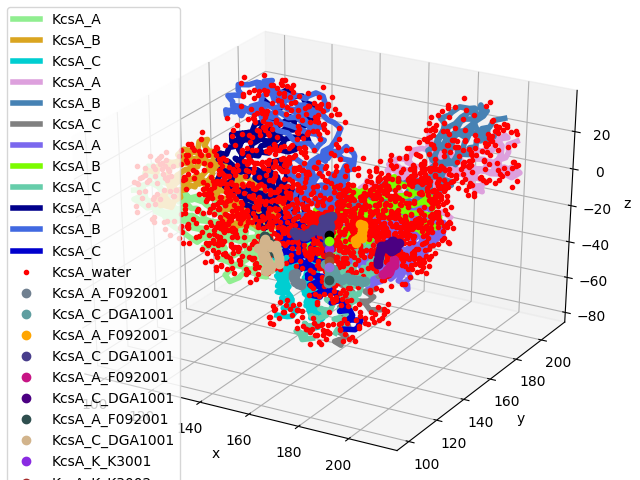
Let’s get a list of all the chains:
In [28]: list(kcsa.iterChains())
Out[28]:
[<Chain: A from Segment A from KcsA (426 residues, 1822 atoms)>,
<Chain: B from Segment A from KcsA (417 residues, 1851 atoms)>,
<Chain: C from Segment A from KcsA (162 residues, 854 atoms)>,
<Chain: A from Segment B from KcsA (426 residues, 1822 atoms)>,
<Chain: B from Segment B from KcsA (417 residues, 1851 atoms)>,
<Chain: C from Segment B from KcsA (162 residues, 854 atoms)>,
<Chain: A from Segment C from KcsA (426 residues, 1822 atoms)>,
<Chain: B from Segment C from KcsA (417 residues, 1851 atoms)>,
<Chain: C from Segment C from KcsA (162 residues, 854 atoms)>,
<Chain: A from Segment D from KcsA (426 residues, 1822 atoms)>,
<Chain: B from Segment D from KcsA (417 residues, 1851 atoms)>,
<Chain: C from Segment D from KcsA (162 residues, 854 atoms)>,
<Chain: K from Segment from KcsA (7 residues, 7 atoms)>]
You see that chain identifiers are preserved within monomers, and monomers have different segment names. To get chain B from first monomer with segment name A, we would do the following:
In [29]: kcsa['A', 'B']
Out[29]: <Chain: B from Segment A from KcsA (417 residues, 1851 atoms)>
