Custom Gamma Functions¶
This example shows how to develop custom force constant functions for
ANM (or GNM) calculations.
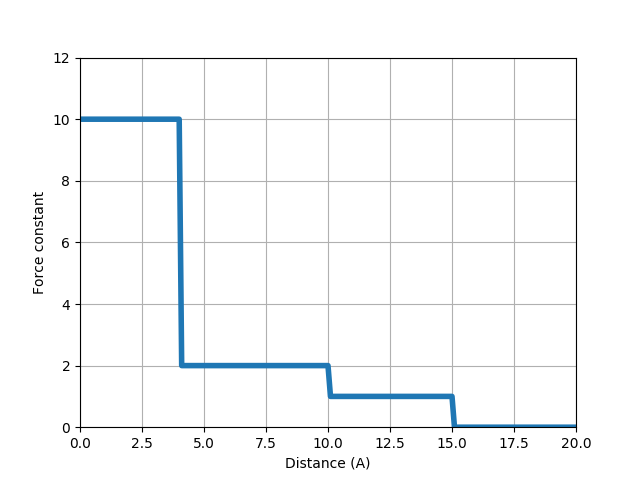
We will use the relation shown in the figure below. For Cα atoms that are 10 to 15 Å apart from each other, we use a unit force constant. For those that are 4 to 10 Å apart, we use a 2 times stronger force constant. For those that are within 4 Å of each other (i.e. those from connected residue pairs), we use a 10 times stronger force constant.
We will obtain an ANM instance that stores Hessian and Kirchhoff
matrices and normal mode data describing the intrinsic dynamics of the protein
structure. ANM instances and individual normal modes
(Mode) can be used as input to functions in prody.dynamics
module.
Parse structure¶
We start by importing everything from ProDy, NumPy, and Matplotlib packages:
In [1]: from prody import *
In [2]: from matplotlib.pylab import *
In [3]: ion() # turn interactive mode on
We start with parsing a PDB file by passing an identifier.
In [4]: p38 = parsePDB('1p38')
In [5]: p38
Out[5]: <AtomGroup: 1p38 (2962 atoms)>
We want to use only Cα atoms, so we select them:
In [6]: calphas = p38.select('protein and name CA')
In [7]: calphas
Out[7]: <Selection: 'protein and name CA' from 1p38 (351 atoms)>
Force Constant Function¶
We define the aformentioned function as follows:
In [8]: def gammaDistanceDependent(dist2, *args):
...: """Return a force constant based on the given square distance."""
...: if dist2 <= 16:
...: return 10
...: elif dist2 <= 100:
...: return 2
...: elif dist2 <= 225:
...: return 1
...: else:
...: return 0
...:
Note that the input to this function from ANM or GNM
is the square of the distance. In addition, node (atom or residue) indices
are passed to this function, that’s why we used *args in the function
definition.
Let’s test how it works:
In [9]: dist = arange(0, 20, 0.1)
In [10]: gamma = map(gammaDistanceDependent, dist ** 2)
In [11]: plot(dist, gamma, lw=4);
In [12]: axis([0, 20, 0, 12]);
In [13]: xlabel('Distance (A)');
In [14]: ylabel('Force constant');
In [15]: grid();
ANM calculations¶
We use selected atoms (351 Cα’s) and gammaDistanceDependent function
for ANM calculations as follows:
In [16]: anm = ANM('1p38')
In [17]: anm.buildHessian(calphas, cutoff=15, gamma=gammaDistanceDependent)
In [18]: anm.calcModes()
For more detailed examples see Anisotropic Network Model (ANM) or Gaussian Network Model (GNM).
